A goal of the NSF-Simons Center for Quantitative Biology is to transform our understanding of organismal growth and development through quantitative approaches. Within the Center, five vibrant research programs are supported in which teams of mathematical and life scientists will closely collaborate. The Center deploys three fundamental mathematical disciplines: dynamical systems theory; stochastic processes; and dimension reduction. These approaches are highly suited to the real-world features of growth and development. Experimental focus will be on established laboratory model organisms such as Drosophila, Caenorhabditis elegans, Xenopus, and mouse.

Project 1: Embryonic Development
During embryonic development, cells progressively and irreversibly become restricted in their potential to become different parts of the adult body. The goal of this project is to discover the mechanisms that generate directed dynamics in development. Mathematical inquisition of cell states within Xenopus embryos and mouse ES cells will be performed. Dynamical systems modeling will connect the small-scale features of the embryonic gene regulatory network to its emergent properties.
Investigators:
Carole LaBonne
Rosemary Braun
Project 2: Growth and Environment
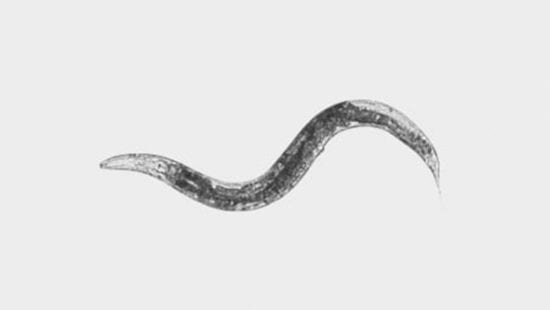
Environmental temperature and diet are two variables that are omnipresent features of life, and both have complex effects on the growth of organisms as they develop. The goal of this project is to provide a rigorous and principled language with which to describe and explore the phenomenon of adaptive responses to environment. The roundworm Caenorhabditis elegans will be used to learn the emergent mathematical rules relating temperature and diet to growth. What are the genomic and metabolic underpinnings of these rules?
Investigators:
Erik Andersen
Niall Mangan
Project 3: Environmental Cycles
Ravi Allada
William Kath
Project 4: Pluripotency
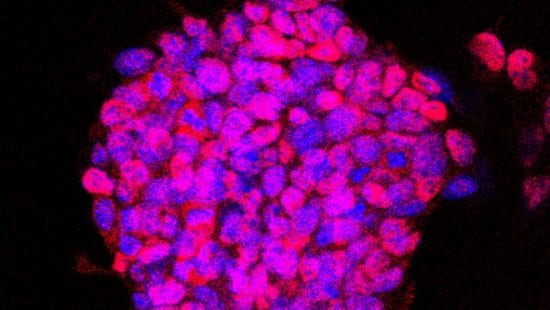
This project conducts single-cell CRISPRi and RNA-seq analysis on mouse pluripotent embryonic cells in culture. The project induces their lineage restriction to neural, mesodermal, and neural crest states using established culture treatments. These data are subject to dimensional reduction in order to discover the gene regulatory network dynamics during maintenance of pluripotency and transition away from pluripotency. This provides insight into how these stages of embryonic development are conserved in terms of cell-state dynamics.
Investigators:
Alec Wang
JiPing Wang

Project 5: Development and Stochasticity
Over time, protein numbers randomly fluctuate for each and every gene within a cell. The goal of this project is to assess the scale of developmental variability that is caused by stochastic processes linked to gene regulation. This will involve exploring the dynamical systems that transform micro-scale molecular stochasticity into macro-scale developmental outcomes in Drosophila. Study will be done on how the topology of the underlying genetic circuits constrains system-level stochastic features.
Investigators:
Richard Carthew


